With the aim of improving the microscopic analysis of digitized histology sections, a CEA/CNRS team from MIRCen (Molecular Imaging Research Center) is forwarding an original approach that combines 3D reconstructions of mouse brain tissue and an analysis based on the resulting voxels, i.e., the discrete digital elements composing the reconstructions. The method was successfully shown to automatically and precisely localize variations in amyloid load, a key aspect of Alzheimer's disease. The study was carried out in partnership with an Inserm team at the Institut Pasteur in Lille and published in Frontiers in Neuroscience (2018).
To validate their approach, the team used murine models of Alzheimer's disease with and without overexpression of the protein ADAM30. Indeed, in a first study1 and using a digital, regionally-segmented mouse brain atlas, the researchers had already observed significantly less amyloid plaques in the hippocampus of mice overexpressing ADAM30. In the new study, with their approach uniting 3D histological reconstruction and voxel-based analysis, the team was able to detect variations in amyloid load automatically and with greater precision within anatomical subregions. They thus confirmed the results of the first study and furthermore identified an additional zone in the cortex where the amyloid load was significantly reduced.
As in the first study, the researchers began by creating 3D reconstructions of histological sections of the mouse brains (those with Alzheimer's disease alone (controls) and those with Alzheimer's disease and ADAM30 overexpression) individually. They then generated an anatomical template by stacking the block-face images and calculating a mean from them. The next step was to position the quantitative map of the amyloid loads in each of the 3D reconstructed brains on the anatomical template. Thereafter, the team was able to perform statistical analyses for the entire brain to automatically produce a detailed map of zones within which amyloid load varied, and in so doing, they greatly enlarged the exploitation of cerebral histological sections.
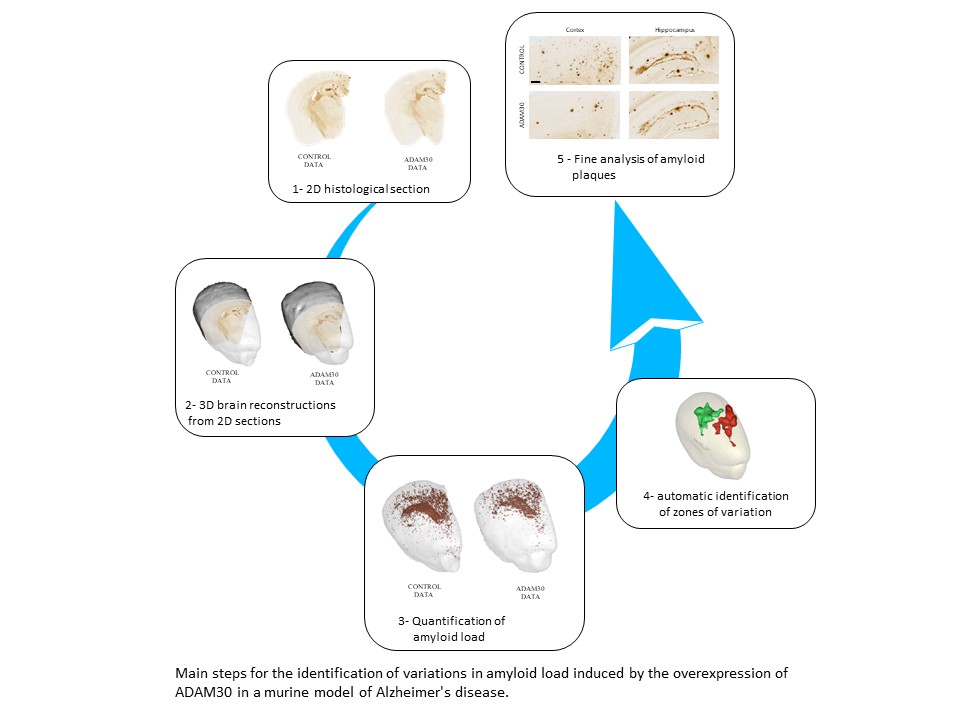
The precise identification of the cerebral zones where amyloid load varies also contributes to a more refined characterization of these amyloid plaques (their size, number, etc.) and a better understanding of their points of origin, evolution and propagation within the brain.
This new, generic approach may also be pertinent for the brain-wide study of complex cellular phenomena, such as neuronal death or neuroinflammation. However, it does require extensive digital resources, both in terms of storage (several terabytes per brain) and calculating power (several thousands of hours of calculations per brain). The laboratory is currently working to adapt and transfer its original approach to the CEA's supercomputers at the TGCC (the "Very Big Center for Calculations") in Bruyères-le-Châtel.
ADAM30 Downregulates APP-Linked Defects Through Cathepsin D Activation in Alzheimer's Disease, Letronne et al., EBiomedicine, 2016